A Synthetic Genetic Edge Detection Program
Jeffrey J. Tabor, Howard Salis, Zachary B. Simpson, Aaron A. Chevalier, Anselm Levskaya, Edward M. Marcotte, Christopher A. Voigt, and Andrew D. Ellington-Department of Pharmaceutical Chemistry, University of California San Francisco, San Francisco,Center for Systems and Synthetic Biology and Institute for Cell and Molecular Biology, Department of Chemistry and Biochemistry, University of Texas, Austin, TX.
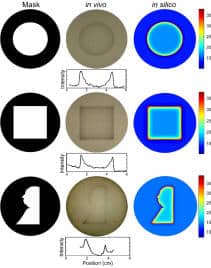
Edge detection is a signal processing algorithm common in artificial intelligence and image recognition programs. We have constructed a genetically encoded edge detection algorithm that programs an isogenic community of E.coli to sense an image of light, communicate to identify the light-dark edges, and visually present the result of the computation. The algorithm is implemented using multiple genetic circuits. An engineered light sensor enables cells to distinguish between light and dark regions. In the dark, cells produce a diffusible chemical signal that diffuses into light regions. Genetic logic gates are used so that only cells that sense light and the diffusible signal produce a positive output. A mathematical model constructed from first principles and parameterized with experimental measurements of the component circuits predicts the performance of the complete program. Quantitatively accurate models will facilitate the engineering of more complex biological behaviors and inform bottom-up studies of natural genetic regulatory networks. The images projected onto the slabs have power characteristics of 0.08 to 0.15W/m2 in the 620-680nm band as determined by a EPP2000C Concave Grating spectrometer (StellarNet, Oldsmar, FL). View PDF…